News
Energy Landscape computed by analyzing single-particle cryo-EM images of ~850,000 ribosomes purified from yeast
Posted by: Joachim Frank |
December 1, 2014 |
No comment
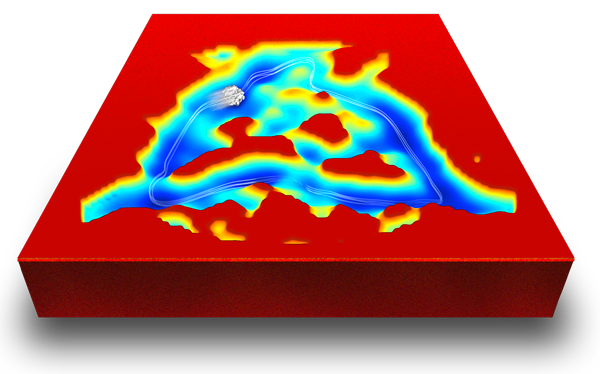
The picture shows the result of a novel method of analysis, by manifold embedding, as applied to a large dataset of ribosomes. These ribosomes are not engaged in translation as they lack mRNA and most tRNAs. However, they still undergo Brownian motions, and the analysis reveals the continuum of conformational changes that thermal bombardment at room temperature will produce.
What we see is a three-dimensional landscape in which the x,y axes represent reaction coordinates (these are orthogonal coordinates associated with the system’s two primary degrees of freedom), and the z coordinate represents the free energy, computed from a 2D array of occupancies (numbers of ribosomes in elements of the x,y grid). This computation follows the Boltzmann relationship; see Fischer et al., Nature 2010.
As customary, the regions of high free energy (or low occupancy) are painted in red, and the regions of low free energy in blue. Therefore, literally, a molecule that is set loose at any point of the landscape will go along a geodesic trajectory, just as a marble would if set loose in a landscape on this planet.
Approximately one third of the ribosomes in the dataset analyzed are in the blue valley, and their trajectory follows the striking closed triangular path. This idea is indicated by the blurred ribosome in motion and the white lines indicating a closed track.
For each point along the trajectory (and indeed, for any point in the landscape), we can collect a bunch of data lying close by, and do a 3D reconstruction just from those. This way, we can find out the conformational changes that the ribosome undergoes when it moves from one place to the other.
The article we just published in PNAS (Dashti et al., 2014) not only describes the new method of analysis, but also contains links to movies that show these conformational changes – intersubunit movements and domain movements. Our analysis of these domain movements confirms an intuitive conclusion: that the triangular blue trajectory represents combinations of motions that are associated with the elongation cycle.
In other words, ribosomes even when they have nothing to do idle away in the same manner as they will do when they are productively engaged in protein synthesis. This realization opens up another dimension of evolution, as acting on ensembles.
Perhaps what needs to be emphasized, since it was not the focus of the PNAS article, is that the new method facilitates the classification of single particle images coming from molecules that exhibit continuous changes of any kind, no matter whether or not they originate from actions of a molecular machine. It is therefore possible now to replace maximum likelihood methods which allow only a limited number of discrete classes to be specified by the manifold embedding approach which contains no such limitation.



