News
A Brief Guide to Three Papers from Our Group Related to Structural Heterogeneity
Posted by: Joachim Frank |
April 3, 2020 |
No comment
Structural heterogeneity has been long recognized as a problem for structure research by cryo-EM. Different molecule species, or molecules in different conformational states or binding configurations may co-exist.
Without resolving this heterogeneity, reconstructions are blurred, and are invalid as representations of any one physical structure. Existing, widely used approaches to resolving heterogeneity, such as Relion, are based on the assumption that the structure exists in a few discrete conformations. Beginning with a paper we published jointly with the group of Abbas Ourmazd at the University of Wisconsin in Milwaukee (1), we have addressed the case of continuous heterogeneity, and how to characterize it by analyzing cryo-EM images of large ensembles of molecules. We are referring to this machine-learning approach as ManifoldEM.
Three recent papers of our group addressing structural heterogeneity of the continuous type (2,3, 4) are worthy of note. They deal with the following issues: Simulation of a heterogeneous cryo-EM dataset for a molecule with continuous conformational variability (2), propagation of conformational coordinates across the angular sphere (3), and navigating energy landscapes (4).
This is a brief note explaining the relationships among these three papers.
While paper (3) is directly related to the ManifoldEM approach (1), and solves one of its long-standing problems, the other two papers, (2) and (4), are quite general, and indeed applicable in the study of structural heterogeneity by any computational method.
SIMULATION OF A HETEROGENIOUS CRYO-EM DATA SET
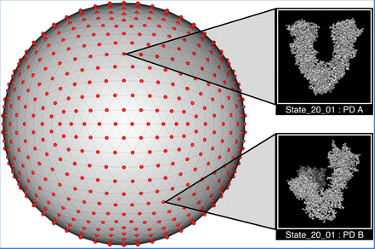
Seitz et al. (2) recognize the need for ground truth data, with which any programs that are addressing with structural heterogeneity can be tested. In this paper, the authors describe a method to generate such data, from a structure in the pdb, with realistic angular distributions, SNR ratios, CTFs, continuous conformational changes with a given distribution of occupancies. They then generated one set of 800,000 single particle images for a particular structure found in the pdb.
PROPAGATION OF CONFORMATIONAL COORDINATES
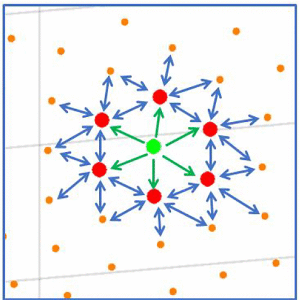
Maji et al. (3) have devised an automated way to propagate conformational coordinates (obtained through Manifold analysis via ManifoldEM) from a projection direction across the whole angular sphere. This part could previously only be done by tedious visual comparisons between successive projection directions, but it is instrumental for deriving a consolidated map of occupancies, from which the energy landscape of the molecule can be derived.
NAVIGATING ENERGY LANDSCAPES
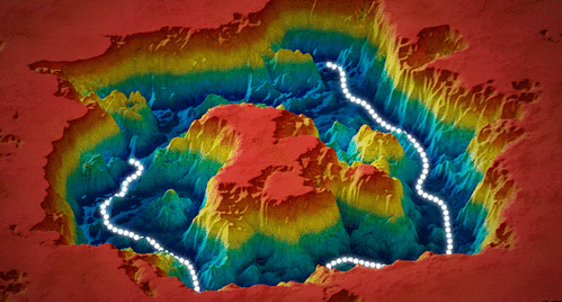
Finally, Seitz and Frank (4) make the assumption that an energy landscape has already been successfully determined (or estimated), using ManifoldEM or any of the other methods recently proposed, and ask the question what pathways of lowest energy a molecule would take in that landscape. They developed a novel method to construct such a pathway.
REFERENCES
(1) Dashti, A., Schwander, P., Langlois, R., Fung, R., Li, W., Hosseinizadeh, A., Liao, H.Y., Pallesen, J., Sharma, G., Stupina, V.A., Simon, A.E., Dinman, J., Frank, J., and Ourmazd, A. (2014). Trajectories of the ribosome as a Brownian nanomachine. Proc Natl. Acad. Sci. USA 111, 17492-17497.
(2) Seitz, E.E., Acosta-Reyes, F., Schwander, P., and Frank, J. (2019) Simulation of cryo-EM ensembles from atomic models of molecules exhibiting continuous conformations. bioRxiv 864116; ALSO: Seitz et al., J. Struct. Biol., submitted.
(3) Maji, S., Liao, H., Dashti, A., Mashayekhi, G., Ourmazd, A., and Frank , J. (2020). Propagation of conformational coordinates across angular space in mapping the continuum of states from cryo-EM data by manifold embedding. J. Chem. Inf. Model.
(4) Seitz, E. and Frank, J. (2020).POLARIS: Path of least action analysis on energy landscapes. J. Chem. Inf. Model.



