News
Just posted in BioRxiv
Posted by: Joachim Frank |
July 26, 2017 |
No comment
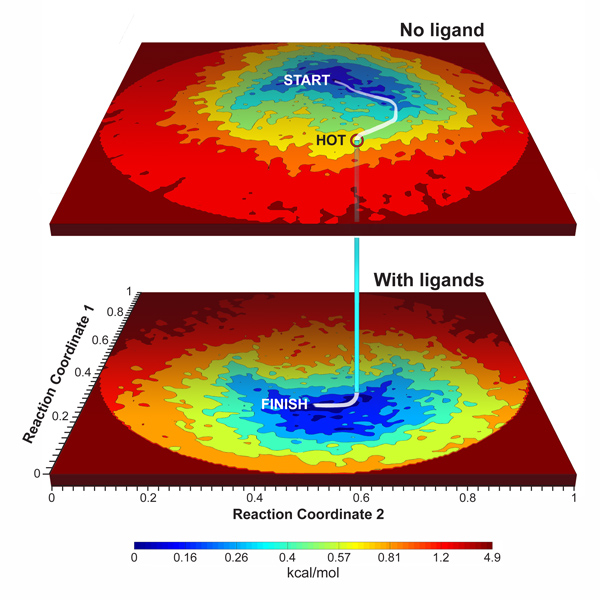
“Conformational Dynamics and Energy Landscapes of Ligand Binding in RyR1”
Ali Dashti, Danya Ben Hail, Ghoncheh Mashayekhi, Peter Schwander, Amedee des Georges, Joachim Frank, Abbas Ourmazd
Click here to read. Biorxiv doi: https://doi.org/10.1101/167080
This study uses the technique of Manifold Embedding, introduced into cryo-EM by an earlier article by Dashti et al. (PNAS 2014), to investigate the mechanism of Ryanodine receptor/Calcium release channel activation and gating. A large number of images (about 400,000 of each) for RyR molecules with and without ligands present were available from the recently published cryo-EM study by Des Georges et al. (Cell 2016). In that study, data were analyzed by standard methods: by using RELION for finding highly populated classes, showing the channel in various open and closed states, modeling structures for each of the cryo-EM maps, and then morphing between these to obtain clues on the mechanisms of ligand binding, channel activation and gating.
With the technique introduced by us and our collaborators in 2014, the data are processed in an entirely different way, leading to a mapping of conformations/states as a (two- or higher-dimensional) continuum. Such occupancy maps can be converted into free-energy landscapes.
Applied to the channel, we were able to trace the trajectory taken by the molecules to assume the conformation in which binding of the ligands, and transition to the bound states is most likely to occur. This “true” trajectory is dissimilar from the trajectory inferred from morphing — a simplistic operation in which the paths of individual atoms are essentially interpolated between two given states.
As the example with the Ryanodine receptor shows, this ensemble analysis of cryo-EM data gives profound insights into the functional dynamics of a biological molecule — insights not available with the standard method of maximum-likelihood classification.



